At Children’s Hospital of Philadelphia (CHOP), you’ll find countless ways to change lives. Our diverse community of more than 22,000 Breakthrough Makers will inspire you to pursue passions, develop expertise, and drive innovation.
Careers at CHOP
Careers at CHOP
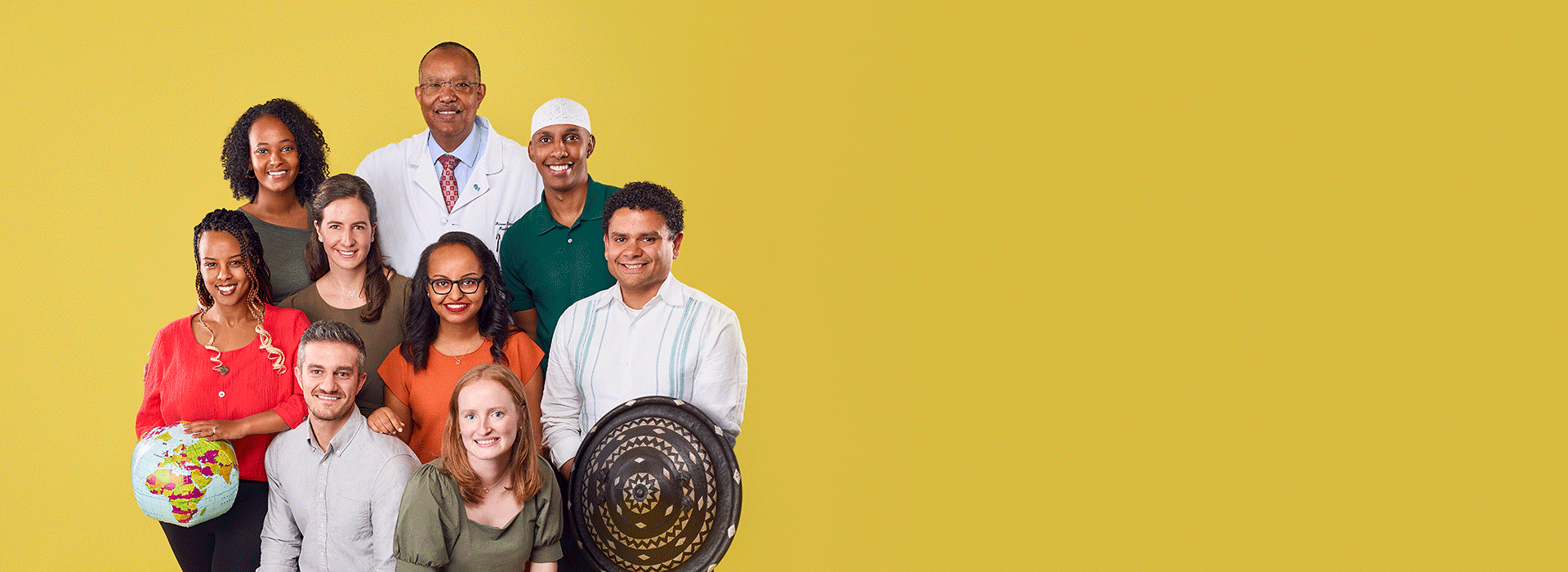
Since 2008, more than 50 CHOP staff have helped grow this partnership and enable its success. There are now five pediatric radiologists out of the 350 practicing in Ethiopia, a country with 120 million people (40% of whom are children).

After confirming her findings with Technical Specialist Marybeth Helfrich, the duo knew there wasn’t time to collect additional samples. With expert knowledge and communication, they worked around this protocol, sent the same sample for definitive testing, and expedited the delivery of lifesaving care.

When a patient showed signs of distress before a routine EKG, social workers and mental health clinicians—who happened to be at the Specialty Care site that day—stepped in to support. They employed multiple techniques, from breathwork to stress balls, to comfort the patient and normalize his feelings.
After, the patient's nurse practitioner and social workers developed a plan for his next visit. Thanks to their compassionate and collaborative efforts, the test was completed in 15 minutes, without incident.
After, the patient's nurse practitioner and social workers developed a plan for his next visit. Thanks to their compassionate and collaborative efforts, the test was completed in 15 minutes, without incident.



